Dipl.-Biol. Mag. Dr. Rose Samuel, ret.

Dipl.-Biol. Mag. Dr. Rose Samuel, ret.
email: mary.rosabella.samuel[@]univie.ac.at
phone: +43 1 4277 54162
room: 219
address: Department of Botany and Biodiversity Research
University of Vienna, Faculty of Life Sciences
Rennweg 14, A-1030 Vienna, Austria
Current Research
- DNA Barcoding and community structure assessment of a tropical forest: a 25 ha mixed dipterocarp forest at Kuala Belalong Brunei Darussalam as a model
- Biodiversity and evolution of New Caledonian Diospyros (Ebenaceae)
- Biogeography and evolution of Nicotiana section Suaveolentes
- Phylogeny and population genomics of peppers from Sri Lanka
DNA Barcoding and community structure assessment of a tropical forest: a 25 ha mixed dipterocarp forest at Kuala Belalong Brunei Darussalam as a model
Project leader: Prof. Rosabelle Samuel
Funded by the Austrian Science Fund (FWF).
Collaborative project with Mark W. Chase (Royal Botanic Gardens, Kew, UK and The University of Western Australia, Australia), Toby Pennington (Royal Botanic Garden, Edinburgh, UK) and Kamariah Abu Salim (University of Brunei Darussalam)
Project members from the University of Vienna: Ovidiu Paun, Jacqueline Heckenhauer, Michael H. J. Barfuss
DNA barcoding is a fast and reliable tool to assess and monitor biodiversity and has a role to play in investigating processes that are responsible for species interactions and thus the community structure of forests. Here, we use DNA barcoding to assess phylogenetic community structure of a 25 ha forest dynamics plot in a mixed dipterocarp forest in Brunei Darussalam which is part of the Center for Tropical Forest Science - Forest Global Earth Observatory (CTFS-ForestGEO) network of plots. Leaves and bark of shrubs and trees were sampled from different subplots (10x10 m) that varied in relative elevation, convexity and slope. We generate DNA sequence data for the two widely used plastid coding regions rbcL and matK for approx. 600 morphotaxa identified in the 70 subplots. These represent 24 orders, 69 families and 189 genera with Dipterocarpaceae (16%) and Euphorbiaceae (9%) as the dominant families. Based on the barcode sequences, we reconstruct phylogenetic relationships among the taxa which will be used to calculate two widely used metrics of PD, Mean Phylogenetic Distance (MPD) and Mean Nearest Taxon Distance (MNTD). Further, Webb’s Net Relatedness index (NRI) and Nearest Taxon Index (NTI) will be calculated. The metrics will be compared to a null model of community assembly to determine whether species are assembled into local communities at random or if they are more closely (phylogenetic clustering) or more distantly (overdispersion) related than expected by chance. Besides biotic interactions habitat filtering often plays a role in species interactions. We will investigate the influence of environmental factors on community structure.
Further, we are interested in the phylogenetic relationships of the South Asian subfamily Dipterocarpoideae which remain poorly resolved, barcoding its species using small fragments of matk and rbcL leads to an unresolved backbone in the phylogenetic tree since closely related species have identical sequences. In order to address this issue, we conducted phylogenetic analysis using plastid DNA sequences. Here we included most genera of the subfamily Dipterocarpoideae from different parts of Asia (Sri Lanka, Thailand, Brunei), as well as individuals of the other subfamilies Monotoideae and Pakaraimaeoideae. The topologies of the trees provide new insights into the genus Shorea of Dipterocarpoideae, which is not monophyletic. We will use RAD sequencing for further investigations of this genus.
For information visit the project website
Biodiversity and evolution of New Caledonian Diospyros (Ebenaceae)
FWF project P22159-B16
Diospyros forms a large genus of woody flowering plants of the family Ebenaceae. New Caledonia is an archipelago in the south-western Pacific and harbours a great range of diverse habitats and a characteristic flora. In total there are 31 Diospyros species found in New Caledonia, of which 30 are endemic. Previous phylogenetic studies of the genus Diospyros based on sequences of plastid markers, showed the New Caledonian species to form three groups. Two of these groups contain only few species (two and, respectively, five), and the majority of species (24) forms the third group (Fig.1). The New Caledonian Diospyros species are morphologically diverse and occupy ecologically different habitats.
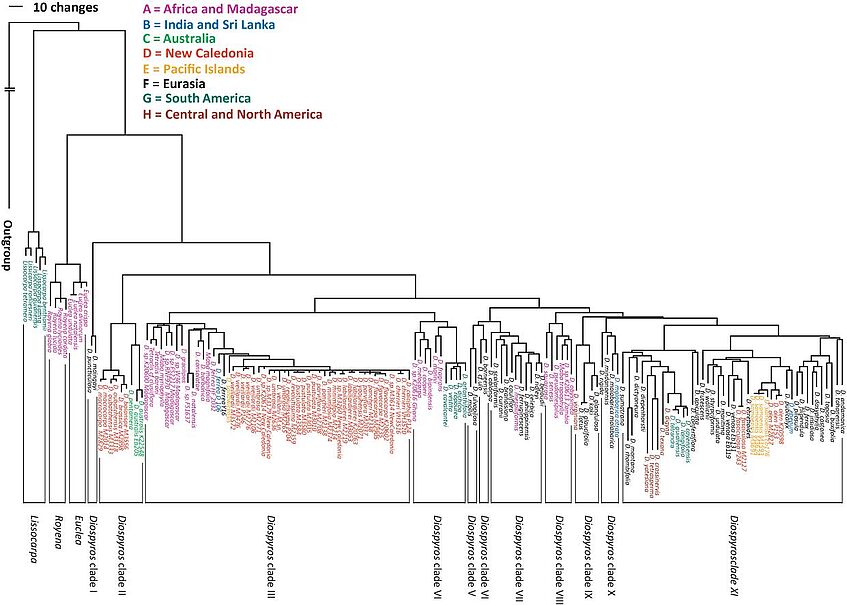
Figure 1. showing the the New Caledonian species marked in red
Sequencing two nuclear and four plastid markers, of the species of group 3 are shows them to be closely related. Diospyros vieillardii is clearly shown to be sister to the rest of this group. Apart from this, individuals of only four species formed unique groups. The morphological species concept, on which the species have been described, is supported by our AFLP results. However, with AFLP data we were not able to elucidate the phylogenetic relationships between these closely related species. Analysis of molecular variance (AMOVA) of the AFLP data set showed that most of the genetic variation occurs within the species rather than among them. Species delimitations inferred from next generation sequencing technique RAD (Restriction site associated DNA) are comparable to those obtained from AFLP data. Phylogenetic trees based on thousands of RAD-derived SNPs are much better resolved than those based on Sanger sequencing of nuclear and plastid markers. Most of the 21 included species formed monophyletic groups in AFLP and RAD analyses. The observed phylogenetic relationships do not follow an ecological structure, pointing to a role of environmental heterogeneity of New Caledonia in shaping speciation events in this group. Functional annotations of genomic regions consistently exhibiting high differentiation between pairs of sister species occurring on different substrates (e.g. D. flavocarpa – D. umbrosa, D. labillardierei – D. trisulca) pointed to genes involved in binding and transporting compounds to/through the cell membrane.
Species from group 3 revealed nearly 3-fold larger genome sizes compared to Diospyros species from other groups (Fig.2a). Chromosome counts showed no indication of polyploidy in this group. The increase in genome size in these species led us to investigate the repeated elements of these genomes.
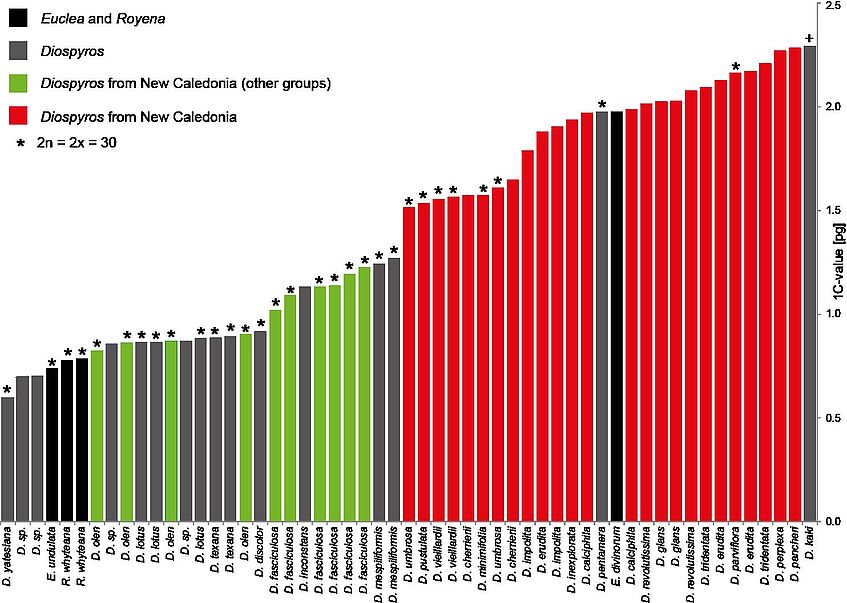
Figure. 2a. Genome size in New Caledonian species
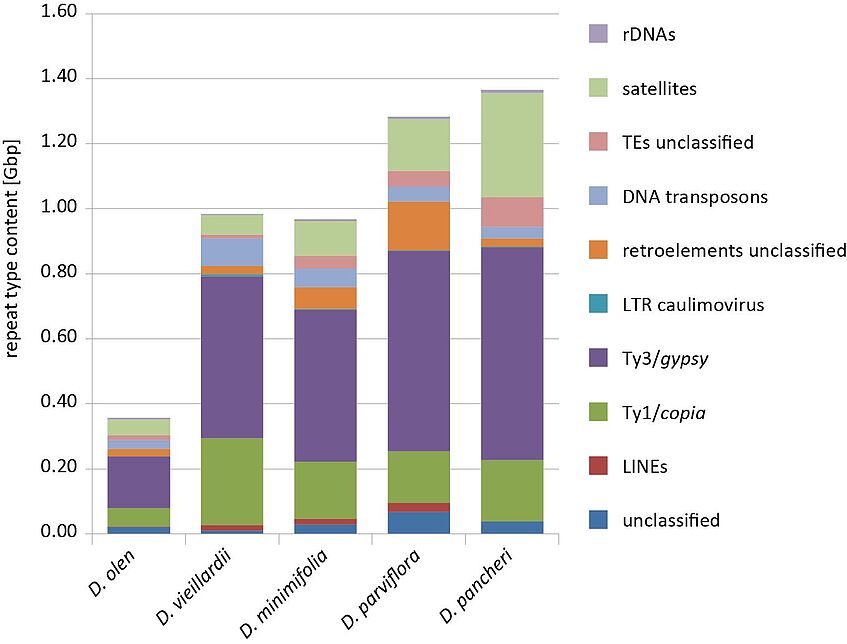
Figure.2b. Copies of the repeated elements present in NC species
Whole genome sequencing using next generation sequencing techniques showed that the larger genomes generally contain more copies of repeated elements such as LTR/gypsy elements, without a significant enrichment for a particular element type (Fig. 2b). Up to now no species specific repeat elements have been identified. Beside the repeated elements we were able to obtain as a by-product whole plastid sequences from the low-coverage whole genome sequencing. The obtained plastomes were compared to the plastid sequence of Camellia sinensis. The plastid genomes of Diospyros and Camellia are highly similar in size, structural organization and gene content. Dating analyses based on DNA sequence and RAD data showed that the crown group 3 is around seven million years old and the group with low statistical support in the RAD based analysis to be around four million years. Diospyros are woody plants with a generation time of several years, thus we can estimate, that not more than 500,000 generations passed since the most recent common ancestor of the latter Diospyros group. The low number of generations after the original long distance dispersal event, together with the rapid radiation across different habitats can explain the presence of the low genetic divergence in this group.
For information visit the project website
Biogeography and Evolution of Nicotiana section Suaveolentes
Project leader: Prof. Rosabelle Samuel
Collaborators: Prof. Dr. Mark W. Chase (Royal Botanic Gardens, Kew, UK), Prof. Ovidiu Paun (University of Vienna), Dr. Luiz Augusto Cauz dos Santos (University of Vienna)
Funded by the Austrian Science Fund (FWF) project number P 33028
Nicotiana section Suaveolentes is widespread in Australia, ranging from a few species in more mesic areas in northern and eastern Australia to the great majority in semi-arid and arid regions, the Eremaean Zone (“from the desert” in Greek) (Figure 1). Hence, the section is particularly species-rich in the central desert around Alice Springs. Nicotiana sect. Suaveolentes appears to be the result of natural allopolyploidy, i.e. hybridisation between individuals belonging to two, not closely related species accompanied by genome doubling. Hence, the allotetraploid ancestor of N. sect. Suaveolentes had 24 pairs of chromosomes. However, the modern representatives of the section exhibit many chromosome numbers, ranging from n = 24–15, and also variation in genome sizes, as we recently described in a paper published in Annals of Botany (Figure 2, Chase et al 2022). The current knowledge of biogeography, phylogenetic relationships and chromosome numbers in N. sect. Suaveolentes are consistent with the ancestral distribution of the genus in Australia being confined to the wetter northern and eastern parts (together with Namibia and the Pacific islands), where all species with higher chromosome numbers (n = 22–24) now occur. Ladiges et al. (2011) believed that the ancestor of N. sect. Suaveolentes is approximately 20 million years old and, like nearly all other Australian plant groups, was formerly widespread across the continent long before severe aridification commenced six million years ago. Two main alternatives exist now in the literature with regard to the timing of radiation of N. sect. Suaveolentes in the Eremaean Zone. The first one is a hypothes that nearly all groups inhabiting this region were already present and had to adapt in parallel to gradual aridification that began six mya. And the alternative scenario suggests that Nicotiana only reached Australia about six mya and started to radiate in the Eremaean Zone just in the last two mya, extending their distributions by dispersal into the extreme habitats that already existed at the time.
In this project we are using RADseq to address the following objectives: (1) clarify species limits in Nicotiana sect. Suaveolentes through species delimitation, (2) clarify phylogenetic relationships so that we can chart their biogeographical patterns, (3) conduct additional molecular clock studies using secondary calibrations (due to no known fossils of Nicotiana) and generation time (with coalescent methods), and (4) investigate the demographics of the group through time, population genetics, population size variation and gene flow. With this additional evidence (5), we will be able to build a stronger case for recentness of the arrival of N. sect. Suaveolentes in Australia and their even more recent expansion into the Eremaean Zone, consistent with scenario in Clarkson et al. (2017, 6 and 2 my, respectively) and in contrast to that in Ladiges et al. (20 my; 2011), which all other evidence to date indicates is much too old. We expect the population genetics studies to reveal patterns consistent with those expected for recently evolved species that achieved their current ranges by dispersal rather than vicariance.
Recent publications from the project:
Cauz-Santos LA, Dodsworth S, Samuel R, Christenhusz MJM, Patel D, Shittu T, Jakob A, Paun O, Chase MW. 2022. Genomic insights into recent species divergence in Nicotiana benthamiana and natural variation in Rdr1 gene controlling viral susceptibility. The Plant Journal in press.
Chase MW, Samuel RM, Leizch AR, Guignard MS, Conran JG, Nollet F, Fletcher P, Jakob A, Cauz-Santos LA, Vignolle G, Dodsworth S, Christenhusz MJM, Buril MT, Paun O. 2022. Down, then up: non-parallel genome size changes and a descending chromosome series in a recent radiation of Australian allotetraploid plant species in Nicotiana section Suaveolentes (Solanaceae). Annals of Botany.
Chase MW, Christenhusz MJM, Palsson RL, Fay MF, Dodsworth S, Conran JG, Cauz-Santos LA, Nollet F, Samuel MR, Paun O. 2021. Species delimitation in Nicotiana sect. Suaveolentes (Solanaceae): reciprocal illumination leads to recognition of many new species. Curti's Botanical Magazine 38: 266-286.
Jakob A. 2021. Phylogenomics of Nicotiana section Suaveolentes (Solanaceae), with a focus on N. benthamiana. MSc Thesis, University of Vienna.

Figure 1: Nicotiana velutina growing in red sand-dune in central South Australia.
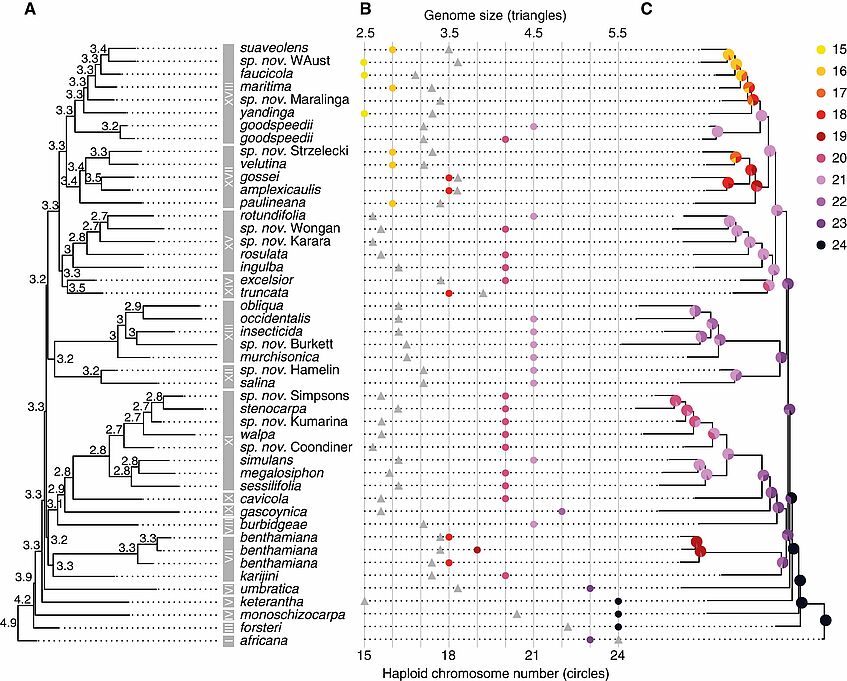
Figure 2: Present state and ancestral reconstruction of chromosome number and genome size. (A) The summary tree of species relationships, as estimated with ChromEvol with chromosome number increases prohibited. (B) Genome sizes (triangles, upper x-scale) and haploid chromosome number (circles, lower X-scale). (C) Genome size evolution as estimated using BayesTree using the summary tree of species relationships. (Chase et al., 2022
Phylogeny and population genomics of peppers from Sri Lanka
Project leader: Prof. Rosabelle Samuel
Collaborators: Prof. Dr. Mark W. Chase (Royal Botanic Gardens, Kew, UK), Prof. Deepthi Yakkandawela (University of Peradeniya), Dr. Subahini Ranasinghe (Royal Botanical Gardens, Peradeniya), Prof. Tara De Silva (University of Colombo), Dr. Anushka Wikramasuriya (University of Colombo), Dr. Luiz Augusto Cauz dos Santos (University of Vienna), MSc. Dominik Metschina (University of Vienna)
Funded by the Austrian Science Fund (FWF) project number P 33143
Piper L. is one of the most diverse genera among the basal clades of angiosperms and widespread in tropical wet forests around the world, having 2,000 species. The monophyly of this large genus has been confirmed by molecular phylogenetic analyses of the Piperales. Molecular analyses done up to now do not include all the Old World species especially those of Sri Lanka. Trimen recorded the occurrence of nine species in Sri Lanka in 1895. A survey done by the Dept. of Export Agriculture of Sri Lanka revealed the presence of eleven species, namely P. nigrum, P. betle, P. longum P.thwaitseii, P.subpeltatum, P argyrophyllum, P. sylvestre, P. zeylanicum, P. trineuron, P. attenuatum and P. chuvya, the last earlier considered a variety of P. betle (Samuel, 1981). The Flora of Ceylon (volume VI, 1987) has recordered 10 species. Of these P. zeylanicum, P. walkerii, P. trineuron are endemics and P. nigrum (black pepper), P. logum (thippili), P. betle (betel leaves) are of economic importance to Sri Lanka.
So far no study has been attempted to examine the phylogenetic relationships of the Sri Lankan species of Piper except for barcoding work (Jayarathna et al., 2016). Molecular phylogenetics have been studied using the nrITS and the plastid intron psbJ-petA, having mainly Neotropical and few Asian and South pacific species (Jaramillo et al., 2008). Here we plan phylogenomic of Sri Lankan Piper species using Next Generation RAD sequencing.
In this project we are using RADseq to address the following objectives: (1) we plan to collect and identify all the wild and cultivated species of the genus Piper, which would contribute to the taxonomic revision of this genus. Morphological characters will be investigated using the herbarium or live specimens of each species (this part of the work will be done by the collaborators, University of Peradeniya, Sri Lanka). (2) we will perform phylogenomic analysis and use coalescent based methods to construct a species tree for Sri Lankan Piper species. This will also permit to test the hypothesis that the endemic Piper species (P. zeylanicum, P. walkerii, P. trineuron) of Sri Lanka have a common ancestor dating up 30-35 mya and their interspecific divergence from the other Sri Lankan wild and cultivated species is shallow. (3) we will perform extensive investigations on the genetic diversity of local selections of P. nigrum (black pepper) available throughout the island (in different agro climatic regions) as well as the introduced cultivars. This will help us to evaluate genetic diversity across the economically important P. nigrum, which will play an important role in crop improvement. (4) we will also use 3D computed tomography to investigate percentage and distribution of bisexual flowers in the spikes of P. nigrum which has an impact on yield of black pepper.

Representatives of all five genera of Piperaceae + subgenus Pleiostachyopiper
Verhuellia lunaria (A), Manekia incurva (B), Zippelia begoniifolia (C), Peperomia pellucida (D), Piper walkeri (E), Piper nudilimbum (F)

Asian Piper species: P. trineuron (A), P. zeylanicum (B), P. kurzii (C), P. griffithii (D), P. porphyrophyllum (E)
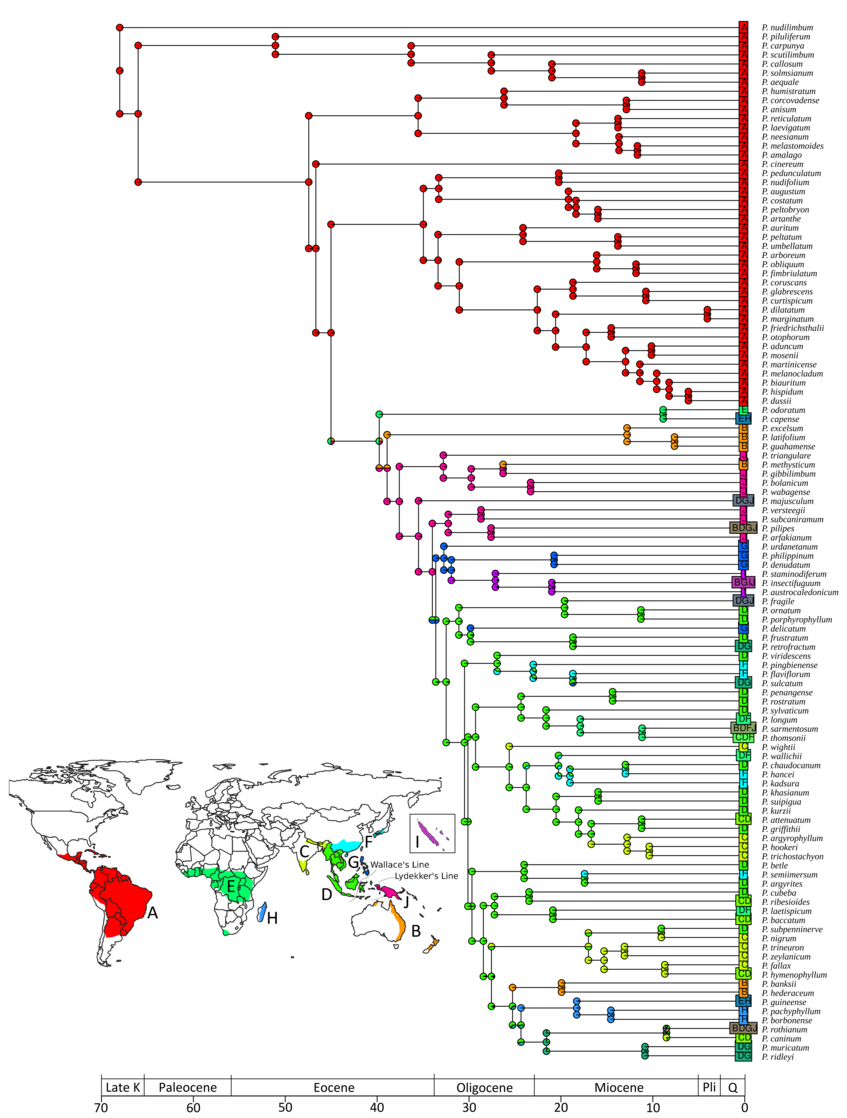
Origin and biogeographical history of Palaeotropical Piper (Piperaceae): multiple long-distance dispersals initiated during the Middle Eocene climatic optimum
Dominik Metschina, Luiz A. Cauz-Santos, Maarten J. M. Christenhusz, James W. Byng, Chalermpol Suwanphakdee, James F. Smith, Imalka Kahandawala, Bhathiya Gopallawa, Nilni Wimalarathna, Anushka M. Wickramasuriya, Michael H. J. Barfuss, Hanna Schneeweiss, Mark W. Chase and Rosabelle Samuel
The ancestral range estimation performed in BioGeoBEARS (BAYAREALIKE+J) estimated the origin of Piper in the Late Cretaceous (70.4 Mya) in South America (Fig. 1). Our investigation of the biogeographical history of Piper via Biogeographical Stochastic Mapping with a coarse range size revealed that out of an average of 141.24 biogeographical events, the majority are within-area speciation events (65.49%). The other ~35% of events are accounted for by founder-event speciation (14.51%) and range expansion (20%), indicating that the occupation of a new area might be a mixture of anagenetic and cladogenetic events. The first founder-event, an intercontinental dispersal of Piper, started during the Middle Eocene climatic optimum (ca. 40 million years ago), first to southern Africa and the Pacific islands, followed by New Guinea, Southeast Asia and the Philippines. In Asia, Piper dispersed and diversified across the Wallace and Lydekker’s Lines multiple times. During the Oligocene, the genus radiated across tropical Asia, dispersing to East Asia, the Himalayas and southern India. Sri Lanka was probably reached from Southeast Asia ca. 28 Mya, where it started diversifying ca. 17 Mya. Africa was reached twice independently, but the genus never diversified there. By the Late Oligocene, Piper had reached its modern distribution.
