Dipl.-Biol. Mag. Dr. Rose Samuel, ret.

Dipl.-Biol. Mag. Dr. Rose Samuel, ret.
email: mary.rosabella.samuel[@]univie.ac.at
phone: +43 1 4277 54162
room: 219
address: Department of Botany and Biodiversity Research
University of Vienna, Faculty of Life Sciences
Rennweg 14, A-1030 Vienna, Austria
DNA Barcoding and community structure assessment of a tropical forest: a 25 ha mixed dipterocarp forest at Kuala Belalong Brunei Darussalam as a model
Project leader: Prof. Rosabelle Samuel
Funded by the Austrian Science Fund (FWF).
Collaborative project with Mark W. Chase (Royal Botanic Gardens, Kew, UK and The University of Western Australia, Australia), Toby Pennington (Royal Botanic Garden, Edinburgh, UK) and Kamariah Abu Salim (University of Brunei Darussalam)
Project members from the University of Vienna: Ovidiu Paun, Jacqueline Heckenhauer, Michael H. J. Barfuss
DNA barcoding is a fast and reliable tool to assess and monitor biodiversity and has a role to play in investigating processes that are responsible for species interactions and thus the community structure of forests. Here, we use DNA barcoding to assess phylogenetic community structure of a 25 ha forest dynamics plot in a mixed dipterocarp forest in Brunei Darussalam which is part of the Center for Tropical Forest Science - Forest Global Earth Observatory (CTFS-ForestGEO) network of plots. Leaves and bark of shrubs and trees were sampled from different subplots (10x10 m) that varied in relative elevation, convexity and slope. We generate DNA sequence data for the two widely used plastid coding regions rbcL and matK for approx. 600 morphotaxa identified in the 70 subplots. These represent 24 orders, 69 families and 189 genera with Dipterocarpaceae (16%) and Euphorbiaceae (9%) as the dominant families. Based on the barcode sequences, we reconstruct phylogenetic relationships among the taxa which will be used to calculate two widely used metrics of PD, Mean Phylogenetic Distance (MPD) and Mean Nearest Taxon Distance (MNTD). Further, Webb’s Net Relatedness index (NRI) and Nearest Taxon Index (NTI) will be calculated. The metrics will be compared to a null model of community assembly to determine whether species are assembled into local communities at random or if they are more closely (phylogenetic clustering) or more distantly (overdispersion) related than expected by chance. Besides biotic interactions habitat filtering often plays a role in species interactions. We will investigate the influence of environmental factors on community structure.
Further, we are interested in the phylogenetic relationships of the South Asian subfamily Dipterocarpoideae which remain poorly resolved, barcoding its species using small fragments of matk and rbcL leads to an unresolved backbone in the phylogenetic tree since closely related species have identical sequences. In order to address this issue, we conducted phylogenetic analysis using plastid DNA sequences. Here we included most genera of the subfamily Dipterocarpoideae from different parts of Asia (Sri Lanka, Thailand, Brunei), as well as individuals of the other subfamilies Monotoideae and Pakaraimaeoideae. The topologies of the trees provide new insights into the genus Shorea of Dipterocarpoideae, which is not monophyletic. We will use RAD sequencing for further investigations of this genus.
For information, visit the project homepage: http://dipterocarpaceae.univie.ac.at
FWF project P22159-B16
Biodiversity and evolution of New Caledonian Diospyros (Ebenaceae)
Diospyros forms a large genus of woody flowering plants of the family Ebenaceae. New Caledonia is an archipelago in the south-western Pacific and harbours a great range of diverse habitats and a characteristic flora. In total there are 31 Diospyros species found in New Caledonia, of which 30 are endemic. Previous phylogenetic studies of the genus Diospyros based on sequences of plastid markers, showed the New Caledonian species to form three groups. Two of these groups contain only few species (two and, respectively, five), and the majority of species (24) forms the third group (Fig.1). The New Caledonian Diospyros species are morphologically diverse and occupy ecologically different habitats.
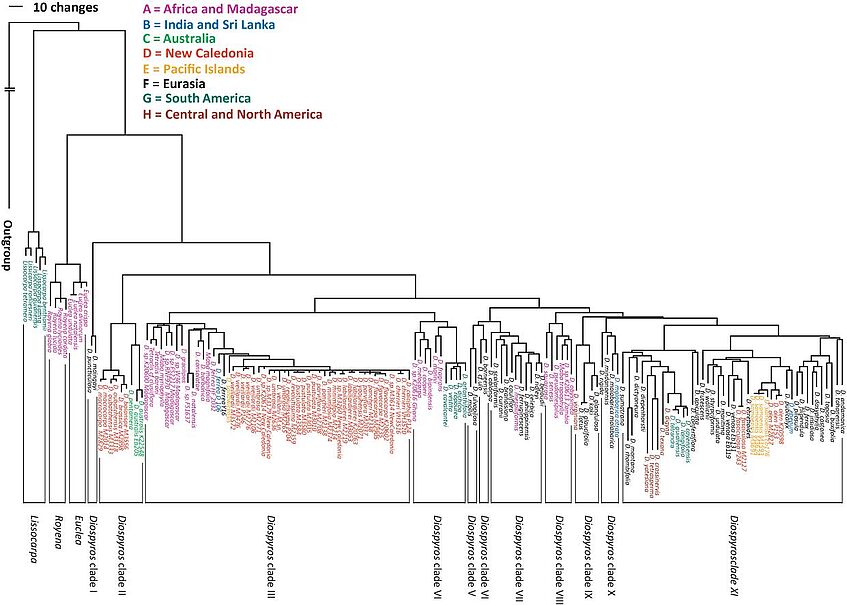
Figure 1. showing the the New Caledonian species marked in red
Sequencing two nuclear and four plastid markers, of the species of group 3 are shows them to be closely related. Diospyros vieillardii is clearly shown to be sister to the rest of this group. Apart from this, individuals of only four species formed unique groups. The morphological species concept, on which the species have been described, is supported by our AFLP results. However, with AFLP data we were not able to elucidate the phylogenetic relationships between these closely related species. Analysis of molecular variance (AMOVA) of the AFLP data set showed that most of the genetic variation occurs within the species rather than among them. Species delimitations inferred from next generation sequencing technique RAD (Restriction site associated DNA) are comparable to those obtained from AFLP data. Phylogenetic trees based on thousands of RAD-derived SNPs are much better resolved than those based on Sanger sequencing of nuclear and plastid markers. Most of the 21 included species formed monophyletic groups in AFLP and RAD analyses. The observed phylogenetic relationships do not follow an ecological structure, pointing to a role of environmental heterogeneity of New Caledonia in shaping speciation events in this group. Functional annotations of genomic regions consistently exhibiting high differentiation between pairs of sister species occurring on different substrates (e.g. D. flavocarpa – D. umbrosa, D. labillardierei – D. trisulca) pointed to genes involved in binding and transporting compounds to/through the cell membrane.
Species from group 3 revealed nearly 3-fold larger genome sizes compared to Diospyros species from other groups (Fig.2a). Chromosome counts showed no indication of polyploidy in this group. The increase in genome size in these species led us to investigate the repeated elements of these genomes.
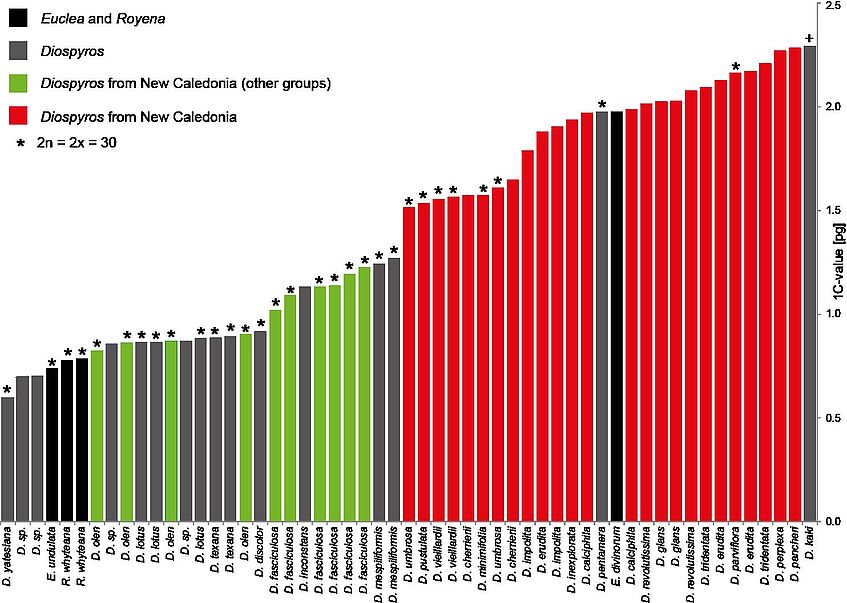
Figure. 2a. Genome size in New Caledonian species
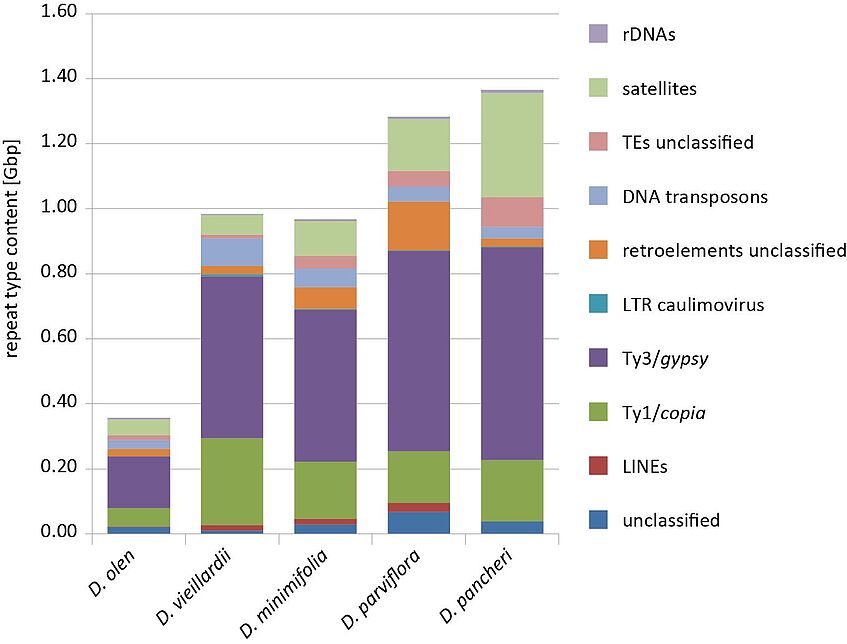
Figure.2b. Copies of the repeated elements present in NC species
Whole genome sequencing using next generation sequencing techniques showed that the larger genomes generally contain more copies of repeated elements such as LTR/gypsy elements, without a significant enrichment for a particular element type (Fig. 2b). Up to now no species specific repeat elements have been identified. Beside the repeated elements we were able to obtain as a by-product whole plastid sequences from the low-coverage whole genome sequencing. The obtained plastomes were compared to the plastid sequence of Camellia sinensis. The plastid genomes of Diospyros and Camellia are highly similar in size, structural organization and gene content. Dating analyses based on DNA sequence and RAD data showed that the crown group 3 is around seven million years old and the group with low statistical support in the RAD based analysis to be around four million years. Diospyros are woody plants with a generation time of several years, thus we can estimate, that not more than 500,000 generations passed since the most recent common ancestor of the latter Diospyros group. The low number of generations after the original long distance dispersal event, together with the rapid radiation across different habitats can explain the presence of the low genetic divergence in this group.
Projectwebsite http://www.botanik.univie.ac.at/systematik/projects/diospyros/index.html
